Joe Felsenstein has posted a fine explanation of how Richard Dawkins’s monkey/Shakespeare model of cumulative selection, better known as the Weasel program, relates to the classic Wright-Fisher model in population genetics. The relation is approximate, because a finite set of offspring is sampled in the monkey/Shakespeare model, while an infinite set of offspring is sampled in the Wright-Fisher model. Joe raised the question of just how well certain results derived for a special case of Wright-Fisher apply to a Weasel-ish evolutionary process. And it happens that I had on hand some code that was easily adapted to address the question. I’m sharing it here, without explanation, and inviting readers to join the discussion at The Skeptical Zone.
A nice illustration of how nice Numeric Python array processing is…
… not that I’m going to explain it.
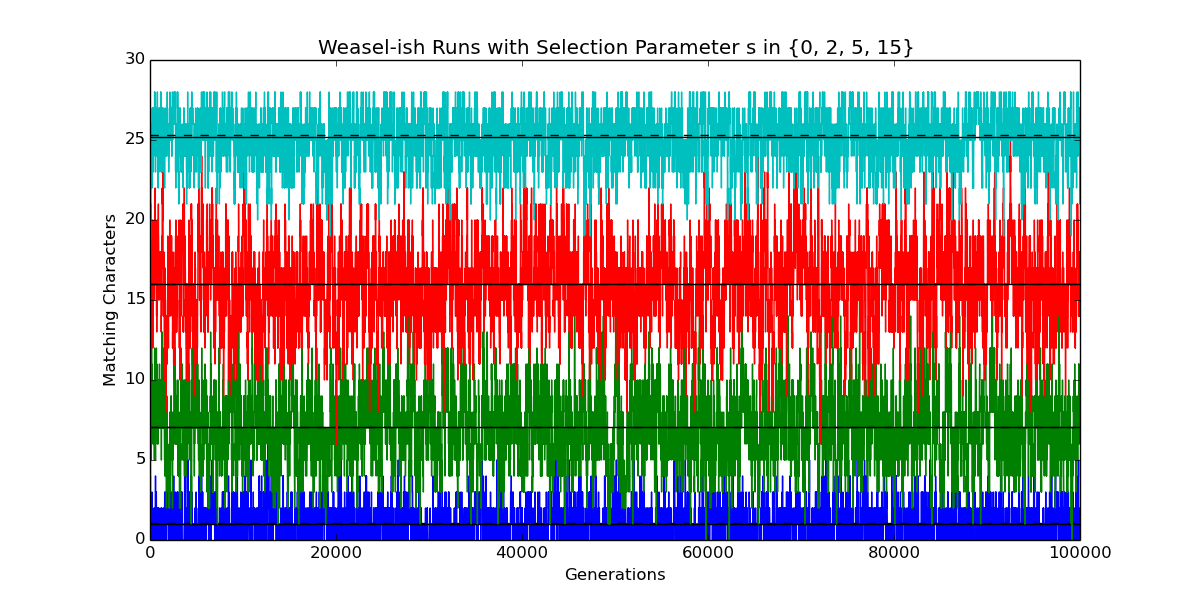
import numpy as np import numpy.random as rand import matplotlib.pyplot as plt LENGTH=28 ALPHABET_SIZE=27 N_OFFSPRING=100 class Evolve(object): """ Model somewhat like Wright-Fisher, with Dawkins's Weasel as a special case. See Felsenstein (2016), "Wright, Fisher, and the Weasel," The Skeptical Zone, http://theskepticalzone.com/wp/wright-fisher-and-the-weasel/. """ def __init__(self, alphabet_size=ALPHABET_SIZE, n_offspring=N_OFFSPRING, mutate_rate=1.0/LENGTH, selection=0.0): """ Initialize evolutionary run. """ self.alphabet_size = alphabet_size self.mutate_rate = mutate_rate self.selection = float(selection) self.probability = np.empty(n_offspring, dtype=float) self.offspring = np.empty(n_offspring, dtype=int) self.n_match = rand.binomial(LENGTH, 1.0 / alphabet_size) self.history = [self.n_match] def generation(self): """ Extend evolutionary run by one generation. Probability of selecting an offspring is proportional to its fitness `(1+selection)**n_match`, where `n_match` is the number of characters matching the target. """ n = len(self.offspring) loss_rate = self.mutate_rate gain_rate = self.mutate_rate / (self.alphabet_size - 1.0) self.offspring[:] = self.n_match self.offspring += rand.binomial(LENGTH - self.n_match, gain_rate, n) self.offspring -= rand.binomial(self.n_match, loss_rate, n) np.power(self.selection + 1.0, self.offspring, self.probability) self.probability /= np.sum(self.probability) self.n_match = rand.choice(self.offspring, size=1, p=self.probability) self.history.append(self.n_match) def extend(self, n_generations=10000): """ Extend evolutionary run by `n_generations` generations. """ for _ in xrange(n_generations): self.generation() def get_history(self, begin=0, end=None): """ Return list of numbers of matching sites for specified generations. """ return self.history[begin:end] def report(self, begin=1000): """ Report on some quantities addressed by Felsenstein (2016). """ u = self.mutate_rate s = self.selection q = u / (1 + s * (1 - u)) p = u * (1 + s) / (self.alphabet_size - 1 + u * s) h = self.history[begin:] print '*'*72 print 'Sentence length L :', LENGTH print 'Alphabet size A :', self.alphabet_size print 'Number of offspring :', len(self.offspring) print 'Mutation rate u :', u print 'Selection parameter s :', s print 'p = u * (1 + s) / (A-1 + u * s) :', p print 'q = u / (1 + s * (1 - u)) :', q print 'Assume equilibrium at generation:', begin print 'Theoretical prob. of match at site p/(p+q) :', p/(p+q) print 'Expected number of matching sites Lp/(p+q) :', LENGTH*p/(p+q) print 'Mean of observed numbers of matching sites :', np.mean(h) print 'Std. dev. of observed nums. of matching sites:', np.std(h) print '*'*72 plt.plot(self.history) m = np.mean(h) plt.plot((0, len(self.history)-1), (m, m), 'k-') m = LENGTH * p/(p+q) plt.plot((0, len(self.history)-1), (m, m), 'k--') def example(): """ Plot numbers of matching sites for runs varying in just one parameter. """ n_gens = 100000 results = [] for s in [0, 2, 5, 15]: e = Evolve(n_offspring=1000, selection=s, alphabet_size=27) e.extend(n_gens) print 'selection (s):', s e.report(begin=5000) results.append(e) plt.title('Weasel-ish Runs with Selection Parameter s in {0, 2, 5, 15}') plt.ylabel('Matching Characters') plt.xlabel('Generations') plt.show() return results results=example() # plt.close()
Clunky, but effective
Here’s the textual output corresponding to the four plots in the figure above, going from bottom to top (blue, green, red, cyan). The idea of this output is to make sure that I’m on the same page with other folks working on the problem.
selection (s): 0 ************************************************************************ Sentence length L : 28 Alphabet size A : 27 Number of offspring : 1000 Mutation rate u : 0.0357142857143 Selection parameter s : 0.0 p = u * (1 + s) / (A-1 + u * s) : 0.00137362637363 q = u / (1 + s * (1 - u)) : 0.0357142857143 Assume equilibrium at generation: 5000 Theoretical prob. of match at site p/(p+q) : 0.037037037037 Expected number of matching sites Lp/(p+q) : 1.03703703704 Mean of observed numbers of matching sites : 1.02184187535 Std. dev. of observed nums. of matching sites: 0.97972454909 ************************************************************************ selection (s): 2 ************************************************************************ Sentence length L : 28 Alphabet size A : 27 Number of offspring : 1000 Mutation rate u : 0.0357142857143 Selection parameter s : 2.0 p = u * (1 + s) / (A-1 + u * s) : 0.0041095890411 q = u / (1 + s * (1 - u)) : 0.0121951219512 Assume equilibrium at generation: 5000 Theoretical prob. of match at site p/(p+q) : 0.252049180328 Expected number of matching sites Lp/(p+q) : 7.05737704918 Mean of observed numbers of matching sites : 7.03611540931 Std. dev. of observed nums. of matching sites: 2.26819646257 ************************************************************************ selection (s): 5 ************************************************************************ Sentence length L : 28 Alphabet size A : 27 Number of offspring : 1000 Mutation rate u : 0.0357142857143 Selection parameter s : 5.0 p = u * (1 + s) / (A-1 + u * s) : 0.00818553888131 q = u / (1 + s * (1 - u)) : 0.00613496932515 Assume equilibrium at generation: 5000 Theoretical prob. of match at site p/(p+q) : 0.571595558153 Expected number of matching sites Lp/(p+q) : 16.0046756283 Mean of observed numbers of matching sites : 15.9726634456 Std. dev. of observed nums. of matching sites: 2.5666185402 ************************************************************************ selection (s): 15 ************************************************************************ Sentence length L : 28 Alphabet size A : 27 Number of offspring : 1000 Mutation rate u : 0.0357142857143 Selection parameter s : 15.0 p = u * (1 + s) / (A-1 + u * s) : 0.021534320323 q = u / (1 + s * (1 - u)) : 0.00230946882217 Assume equilibrium at generation: 5000 Theoretical prob. of match at site p/(p+q) : 0.903141702516 Expected number of matching sites Lp/(p+q) : 25.2879676704 Mean of observed numbers of matching sites : 25.1902401027 Std. dev. of observed nums. of matching sites: 1.54315605955 ************************************************************************
No comments :
Post a Comment